- Next Gen Sequencing (NGS) Myeloid Malignancy Targeted Panel
- Confirmation of a Research Finding
- CYP2C19 Genotyping
- Cytogenomic Microarray
- Extract and Hold
- FLT3 ITD and TKD Fragment Size Analysis
- IDH1 R132 and IDH2 R140, R172 Sanger Sequencing
Next Gen Sequencing (NGS) Myeloid Malignancy Targeted Panel
CPT code: 81455
The detection of gene mutations in hematologic malignancy samples is increasingly important owing to the volume of research demonstrating that many of these variants have an impact on cancer diagnosis, prognosis, and response to treatment. Cancer treatment decisions are increasingly made based on the patient’s genomic information. Genomic tests designed to facilitate decisions about treatment management are based on the ability of the test to accurately and reproducibly identify variants in a single gene or by scanning for mutations across many genes simultaneously. These multi-gene panels include targeted gene-expression profiling tests that are used to estimate prognosis and/or the likelihood of recurrence.
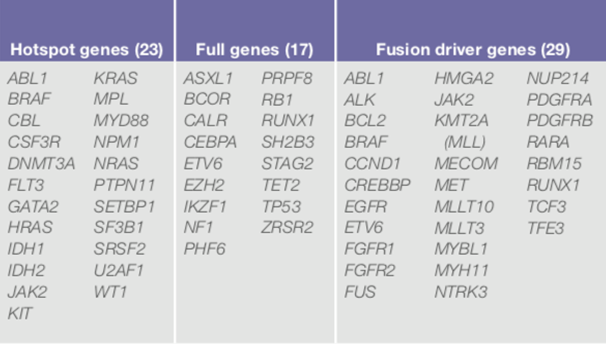
Utilizing the ThermoFisher Oncomine Myeloid™ Assay (OMA), designed for use on the Ion Torrent Chef workstation to automate library preparation, templating, and chip loading. The massive parallel sequencing will be performed on our Ion S5 sequencer for the detection of myeloid malignancies. The panel is comprised of 40 key DNA genes and a broad fusion panel of 29 driver genes (total of 69 genes) to cover the most relevant targets in major myeloid disorders: acute myeloid leukemia (AML), myeloid dysplastic syndrome (MDS), myeloproliferative neoplasms (MPN), chronic myeloid leukemia (CML), chronic myelomonocytic leukemia (CMML), and juvenile myelomonocytic leukemia (JMML).
Multi-gene amplicon-based panels also include DNA and RNA analysis through next-generation sequencing (NGS) technologies, including custom panels that profile multiple actionable driver genes and tumor characteristics that may guide the selection of targeted therapies based on druggable targets.
The panel contains known clinically significant or disease-causing variants in 11 high-priority genes specifically requested by UMMC oncologists: CALR, CEBPA, DNMT3A, FLT3, IDH1, IDH2, JAK2, KIT, MPL, NPM1, and TP53 for the following hematological malignancy types; AML, MDS and MPN.
TGL will not include in this report FLT3-ITD or SNP variants with the coding regions of IDH1 R132 or IDH2 R140 and R172. These variants are reported out separately and more rapidly using existing TGL methodologies including Sanger-based sequencing and fragment size analysis.
Confirmation of a Research Finding
CPT code: 81479
Prior to being reported to physicians and entered into the patient's medical record, sequencing variants identified under research protocols require confirmation by Sanger sequencing in a CLIA-certified laboratory, such as TGL. Confirmation testing under CLIA allows patients and their physicians to receive the results and to use them in clinical management decisions.
An appropriate exon of the gene of interest is analyzed by bidirectional Sanger DNA sequencing from whole blood or bone marrow sample using Sequencer sequencing analysis software. Sequences are compared to the appropriate NCBI reference sequences for the gene of interest. NOTE: TGL must receive an original blood sample from an approved phlebotomy location. Samples extracted by research laboratories will not be accepted for Confirmation of a Research Finding.
CYP2C19 Genotyping
CPT code: 81225
Clopidogrel (Plavix) response is due in part to variation in the Cytochrome P450 2C19 gene (CYP2C19). Six variants (*2,*3, *4, *6, *8, and *17) in the CYP2C19 gene are analyzed by Taqman genotyping for rs121913499.
This assay does not detect other CYP2C19 variants
Cytogenomic Microarray
CPT code: 81229
Genomic imbalance is a major cause of congenital or developmental abnormalities. Genome-wide DNA copy number analysis is performed using the Affymetrix Cytoscan High Density (HD) ™ protocol along with the Command Console and Chromosome Analysis Suite software (CHAS).
This array includes >1.8 million Copy Number Variant probes and >750,000 SNP probes, providing genotyping information that enables detection of copy number neutral loss of heterozygosity (CN-LOH) and detection of some Uniparental Disomy (UPD) cases.
Extract and Hold
NADNA can be extracted in our CLIA/CAP laboratory to allow for an initial work-up to be completed prior to determining if additional testing is necessary. The intent is for DNA to be used for future clinical testing and is not intended for long-term storage or banking of samples. DNA will be extracted by either the EZ1 DNA Blood or the EZ1 DNA Tissue kit and stored for up to twelve months from receipt of the sample.
FLT3 ITD and TKD Analysis
CPT codes: 81245 and 81246
FLT3 is a receptor tyrosine kinase with important roles in hematopoietic stem/progenitor cell survival and proliferation. This assay is used in testing for common FLT3 variants in patients with acute myeloid leukemia (AML). The FLT3 internal tandem duplication (FLT3-ITD) variant is present in about 30 percent of newly diagnosed AML patients and results in a poor prognosis and decreased disease-free survival.
1-5 The FLT3 tyrosine kinase domain (FLT3-TKD) variant occurs in residue D835 or I836 of the kinase domain and confers resistance to FLT3 inhibitors. Fragment size analysis using capillary electrophoresis and the GeneMapper software package of two different regions of the FLT3 gene. Amplification of a 328 base pair (bp) region within the FLT3 gene is achieved using site-specific fluorescently labeled primers. The presence of the FLT3-ITD variant is detected by observing the amplification of this targeted region. A fragment size identical to the negative control is indicative of a negative result and a fragment size larger than the negative control is indicative of a positive result.
The detection of variants within the FLT3-TKD is accomplished by amplifying a region of the DNA coding for this region of the FLT3 protein using fluorescently labeled primers. This product is then digested with a restriction enzyme, EcoRV. Bases coding for the D835 and the I836 codons are included in the restriction site for EcoRV, and variants in these codons will result in loss of the EcoRV site. In the negative control sample, two fragments will be observed as a result of cleavage by the enzyme and if the site is mutated only one larger fragment will be observed.
IDH1 R132_IDH2 R140 and R172 Sequencing
CPT codes: 81120 and 81121
IDH1 (isocitrate dehydrogenase 1 (NADP+), soluble) and IDH2 (isocitrate dehydrogenase 1 (NADP+), mitochondrial) are mutated in approximately 15 - 23 percent of de novo Acute Myeloid Leukemia (AML) cases. Variants have also been identified in 5 percent of myelodysplastic syndromes (MDS) and 10 percent of myeloproliferative neoplasms (MPN).
IDH1 codon 132, IDH2 codons 140 and 172 and the surrounding sequences within exon 4 are analyzed by bidirectional Sanger DNA sequencing for whole blood or bone marrow samples, using Sequencher sequencing analysis software. Sequences are compared to NCBI reference sequences for IDH1 (NM_005896.3 and NP_005887.2) and IDH2 (NM_002168.3 and NP_002159.2) genes. Once a specific variant has been identified within a family, site-specific testing by Sanger sequencing can be performed for other family members, rather than full gene sequencing. An appropriate exon or fragment of the gene of interest is analyzed by bidirectional Sanger DNA sequencing from whole blood or bone marrow sample using Sequencer sequencing analysis software. Sequences are compared to the appropriate NCBI reference sequences for the gene of interest.