CONTACT:

Robert J. Bloch, PhD
Core Director
Department of Physiology
rbloch@som.umaryland.edu

Yinghua Zhang, PhD
Core Operator
Department of Physiology
yinghuazhang@som.umaryland.edu
LOCATION:
Room 611, Health Sciences Facility II
20 Penn Street Baltimore, MD 21201
HOURS:
Monday through Friday
7:00am - 5:00pm
PHONE:
410-706-3020 or
410-706-2036
MISSION:
The Biosensor Core Facility’s objective is to provide the faculty, staff and students on the University of Maryland, Baltimore, with the latest technology for the quantitative study of binding reactions in real time, specifically with an approach that is versatile, highly sensitive, and “user friendly,” with molecules that are label-free. The instruments we use for this purpose are from Biacore® (GE Healthcare).
SERVICES:
Biacore® instruments utilize the optical method of “surface plasmon resonance” (SPR), small changes in the interaction of monochromatic light with a metallic surface that occur when a protein or other molecule binds to that surface. Using the T200 or 3000, the core and its staff can provide accurate determinations of “on” and “off” rates for binding reactions, as well as determine affinity constants for binding. Because our instruments use SPR, many different kinds of binding reactions can be studied, often robotically, and a wide range of biological molecules can be examined, including proteins, nucleic acids, carbohydrates and lipids, as well as small molecules. Typical studies can:
- Determine if pairs of molecules bind to each other.
- Determine kinetic constants, binding constants, and specificity of binding.
- Determine if several molecules can bind simultaneously to the same ligand or if they compete for binding.
INSTRUMENTATION:
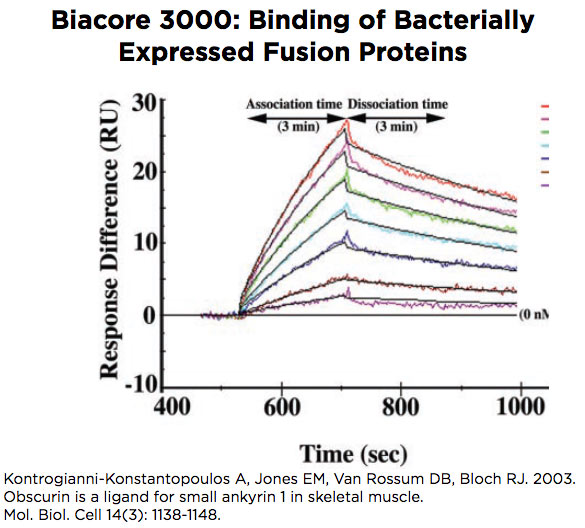
 Biacore 3000
Biacore 3000
The Biacore 3000 is designed to study binding of macromolecules to each other, with the possibility of examining molecules as small as ~2 kDa. The instrument accepts a chip with 4 flow cells that can be used in pairs, to compare flow cell 2 with flow cell 1 and flow cell 4 with flow cell 3, or in a single set of 4, to compare flow cells 2, 3 and 4 each with flow cell 1. Software is designed to optimize curve fitting and calculation of kinetic and binding constants.
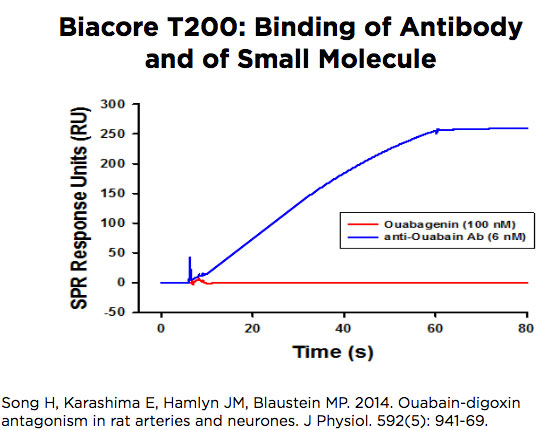
 Biacore T200
Biacore T200
The Biacore T200 operates very similarly to the 3000 but it has a very stable baseline signal which allows it to be used to study the binding of small molecules as well as macromolecules. The instrument accepts a chip with 4 flow cells that can be used in pairs, to compare flow cell 2 with flow cell 1, or flow cell 4 with flow cell 3. The software has been adapted to facilitate kinetics studies in a single cycle, by introducing low to high concentrations over the surface of the chip without intervening wash or regeneration steps.