Academic Title:
Professor
Primary Appointment:
Pharmacology & Physiology
Administrative Title:
Director of the Confocal Microscopy Core; Vice Chair -- Department of Pharmacology & Physiology
Location:
Lab: 5-014 Bressler Research Bldg, 655 W Baltimore St., Baltimore, MD 21201
Phone (Primary):
410-706-4769
Fax:
410-706-8341
Education and Training
I graduated from Yale University with a bachelor's degree in Psychology. My long-standing interest in cognition and learning has lead to my current work to understand the cellular processes that underlie mental health and psychiatric disorder. At the University of Pittsburgh, I obtained a Ph.D. in the Department of Neuroscience with Jon Johnson, Ph.D., where I used single-channel recordings to study the mechanisms by which the anti-Parkinsonian and anti-Alzheimer's drugs amantadine and memantine act on NMDA receptors. I then undertook postdoctoral training with George Augustine, Ph.D. and Michael Ehlers, M.D. Ph.D. at Duke University in the Department of Neurobiology. I joined the Department as an Assistant Professor in 2005, and became Vice-Chair in 2022.
Research/Clinical Keywords
Synaptic transmission, neural plasticity, neuronal cell biology, optical physiology, spatial proteomics, neurophysiology, live-cell confocal imaging, super-resolution imaging, DNA PAINT, single-molecule tracking.
Highlighted Publications
Sarkar D*, Kang J*, Wassie AT*, Schroeder ME, Peng Z, Tarr TB, Tang AH, Niederst E, Young JZ, Su H, Park D, Yin P, Tsai L-H, Blanpied TA, Boyden ES. (2022) Revealing nanostructures in brain tissue via protein decrowding by iterative expansion microscopy. Nature Biomed Eng, 6(9):1057-1073. doi: 10.1038/s41551-022-00912-3 (*co-first authors)
Ramsey AM*, Tang AH*, LeGates TA, Guo X-Z, Carbonne EA, Thompson SM, Biederer T, Blanpied TA. (2021) Subsynaptic positioning of AMPARs by LRRTM2 controls synaptic strength. Science Advances, 7(34):eabf3126 (*co-first authors)
Held RG*, Liu C,* Ma K*, Ramsey AM, Tarr TB, De Nola G, Wang SSH, Wang J, Arn M.J.M. van den Maagdenberg AM, Schneider T, Sun J, Blanpied TA, Kaeser PS (2020) Synapse assembly in the absence of presynaptic Ca2+ channels and Ca2+ Neuron, 107:667-683.
Metzbower SR, Joo Y, Benavides DS, Blanpied TA (2019) Properties of individual hippocampal synapses influencing NMDA-receptor activation by spontaneous neurotransmission. eNeuro, 6(3): e0419-18.
Divakaruni SS, Van Dyke AM, Chandra R, LeGates TA, Contreras M, Dharmasri PA, Higgs HN, Lobo MK, Thompson SM, Blanpied TA. (2018) Long-Term Potentiation Requires a Rapid Burst of Dendritic Mitochondrial Fission during Induction. Neuron 100: 860-875.
Biederer T, Kaeser PS, Blanpied TA. (2017) Transcellular Nanoalignment of Synaptic Function. Neuron. 96:680-696.
Francis TC, Chandra R, Gaynor A, Konkalmatt P, Metzbower SR, Evans B, Engeln M, Blanpied TA, Lobo MK. (2017) Molecular basis of dendritic atrophy and activity in stress susceptibility. Mol Psychiatry. 22:1512-1519.
Tang AH*, Chen H*, Li TP, Metzbower SR, MacGillavry HD, Blanpied TA. (2016) A trans-synaptic nanocolumn aligns neurotransmitter release to receptors. Nature 535 (7215) August 11. (*These authors contributed equally.)
Li TP and Blanpied TA. (2016) Control of Transmembrane Protein Diffusion within the Postsynaptic Density Assessed by Simultaneous Single-Molecule Tracking and Localization Microscopy. Frontiers in Synaptic Neuroscience 8: 8-22-16.
Perez de Arce K, Schrod N, Metzbower SR, Kong G K-W, Tang A, Krupp AJ, Stein V, Liu X, Blanpied TA, Lucic V and Biederer T. (2015) Topographic Mapping of the Synaptic Cleft into Adhesive Nanodomains. Neuron 88(6): 1165–1172.
Lu HE, MacGillavry HD, Frost NA, Blanpied TA (2014) Multiple spatial and kinetic subpopulations of CaMKII in spines and dendrites as resolved by single-molecule tracking PALM. J Neurosci. 2014 May 28;34(22):7600-10
MacGillavry, HD, Song, Y, Raghavachari, S, and Blanpied TA. (2013) Nanoscale scaffolding domains within the postsynaptic density concentrate synaptic AMPA receptors. Neuron. 78: 615-22.
Additional Publication Citations
Dean CA, Metzbower SR, Dessain SK, Blanpied TA, Benavides DR (2022) Regulation of NMDA Receptor Signaling at Single Synapses by Human Anti-NMDA Receptor Antibodies. Front Mol Neurosci. Jul 28;15:940005.
Richard EM, Polla DL, Assir MZ, Contreras M, Shahzad M, Khan AA, Razzaq A, Akram J, Tarar MN, Blanpied TA, Ahmed ZM, Jamra RA, Wieczorek D, van Bokhoven H, Riazuddin S, Riazuddin S. (2019) Biallelic Variants in METTL5 Cause Autosomal Recessive Intellectual Disability and Microcephaly. Am J Hum Gen, 105: 869-78.
Scheefhals N, Westerveld ML, Blanpied TA, Hoogenraad CC, MacGillavry HD (2019) Shank proteins couple the endocytic zone to the postsynaptic density to control trafficking and signaling of metabotropic glutamate receptor 5. Cell Rep. 29: 258-69.
Chen J-H, Blanpied TA, Tang A-H (2020) Quantitative Analysis of Trans-synaptic Protein Alignment: A data analysis case for single-molecule localization microscopy. Methods, S1046-2023(18)30430-4.
Chen H, Tang AH, Blanpied TA. (2018) Subsynaptic spatial organization as a regulator of synaptic strength and plasticity. Curr Opin Neurobiol. 51:147-153.
Li TP, Song Y, MacGillavry HD, Blanpied TA*, Raghavachari S*. (2016) Protein Crowding within the Postsynaptic Density Can Impede the Escape of Membrane Proteins. J Neurosci 36(15):4276-95. (co-corresponding authors)
Kim K, Lakhanpal G, Lu HE, Khan M, Suzuki A, Kato-Hayashi M, Narayanan R, Luyben TT, Matsuda T, Nagai T, Blanpied TA, Hayashi Y, Okamoto K. (2015) A Temporary Gating of Actin Remodeling during Synaptic Plasticity Consists of the Interplay between the Kinase and Structural Functions of CaMKII. Neuron 87(4): 813-26.
Yu D, Makkar G, Strickland DK, Blanpied TA, Stumpo DJ, Blackshear PJ, Sarkar R, Monahan TS. (2015) Myristoylated Alanine-Rich Protein Kinase Substrate (MARCKS) Regulates Small GTPase Rac1 and Cdc42 Activity and Is a Critical Mediator of Vascular Smooth Muscle Cell Migration in Intimal Hyperplasia Formation. J Am Heart Assoc. 2015 Oct 8;4(10). pii: e002255
Jensen, C.S., Watanabe, S., Rasmussen, H.B., Schmitt, N, Olesen, S-P., Frost, N.A., Blanpied, T.A. and Misonou, H. (2014) Specific sorting and post-Golgi trafficking of dendritic potassium channels in living neurons. J Biol Chem 289:10566-81.
Valenzuela JI, Jaureguiberry-Bravo M, Salas DA, Ramírez OA, Cornejo VH, Lu HE, Blanpied T.A., and Couve A (2014 )Transport along the dendritic endoplasmic reticulum defines the trafficking modality for GABAB receptors. J Cell Sci127:3382-95.
MacGillavry HD, Blanpied TA (2013) Single-Molecule Tracking Photoactivated Localization Microscopy to Map Nano-Scale Structure and Dynamics in Living Spines. Curr Protoc Neurosci. 2013;65:2.20.1-19
Kerr, J.M. and Blanpied T.A. (2012) Subsynaptic AMPA receptor distribution is acutely regulated by actin-driven reorganization of the postsynaptic density. J. Neurosci. 32: 658-73.
Frost NA, Lu HE, Blanpied T.A. (2012) Optimization of cell morphology measurement via single-molecule tracking PALM. PLoS One 7(5):e36751
Lieberman JA, Frost NA, Hoppert M, Fernandes PJ, Vogt SL, Raivio TL, Blanpied T.A., Donnenberg MS (2012) Outer Membrane Targeting, Ultrastructure, and Single Molecule Localization of the Enteropathogenic Escherichia coli Type IV Pilus Secretin BfpB. J Bacteriol. 194: 1646-58.
MacGillavry, H.D., Kerr, J.M., Blanpied T.A. (2011) Lateral organization of the postsynaptic density. Mol Cell Neurosci., 48: 321-31.
Weinman E.J., Steplock D, Shenolikar S, Blanpied T. A. (2011) Dynamics of PTH-induced disassembly of Npt2a/NHERF-1 complexes in living OK cells. Am J Physiol Renal Physiol. 300:231-5.
Frost N. A., Kerr J. M., Lu H. E., Blanpied T.A. (2010) A network of networks: Cytoskeletal control of compartmentalized function within dendritic spines. Curr. Op. Neurobiol., 20: 578-87.
Frost N. A., Shroff H., Kong H., Betzig E., Blanpied T. A. (2010) Single-molecule discrimination of discrete perisynaptic and distributed sites of actin filament assembly within dendritic spines. Neuron, 67: 80-90.
Research Interests
Research in my lab examines protein trafficking mechanisms at synapses, and seeks to understand how these mechanisms are used to regulate synaptic transmission.
The dendrites of neurons receive and integrate inputs from hundreds or thousands of partner cells, and most of this input arrives at highly specialized sites called synapses that are distributed over the dendritic tree. Improper regulation of synaptic transmission is implicated in an enormous variety of psychiatric disorders: an imbalance of glutamatergic neurotransmission in particular has been identified in the pathophysiology of diseases ranging from schizophrenia and autism to epilepsy and addiction, and increasing evidence suggests that excitatory synapses are among the earliest targets of Alzheimer's Disease (AD) pathogenesis. A key means of synaptic regulation is the control of the number and subsynaptic position of postsynaptic neurotransmitter receptors, and so understanding the mechanisms involved has broad implications not only for understanding the etiology of many diseases but more generally for defining the cellular basis of nervous system function and disorder.
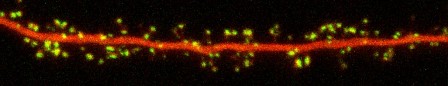
We focus on processes that operate rapidly and very locally to regulate synaptic function on a moment-to-moment basis. These mechanisms are of particular interest because they likely underlie the initial stages of memory formation, and their perturbation produces severe and long-lasting damage to synaptic and neuronal morphology. Mechanisms we are studying currently include the trans-synaptic molecular bridges that align presynaptic nerve terminals with postsynaptic specializations, the continuous internalization and recycling of synaptic receptors, and the postsynaptic actin cytoskeleton that controls postsynaptic morphology and regulates receptor trafficking in many ways.

Understanding the complex set of molecular reactions that underlie endocytosis and other trafficking events requires examining these events in live cells. Thus, the lab has turned to high-resolution, high-speed, multi-color confocal imaging of proteins in cultured neurons as a way of examining the kinetics and spatial features of the molecular events at individual living synapses. We utilize a number of novel fluorescence assays to examine protein positioning and movement at the synapse in living cells at the highest resolution possible. These tools provide the perfect complement to whole-cell and single-channel electrophysiological assays, which naturally track receptor function at high speed but typically with little or no spatial resolution. The lab aims to combine these approaches with molecular perturbation of protein function to provide unprecedented spatial, temporal, and molecular understanding of the protein trafficking events in spines.
Lab Techniques and Equipment
Most research in the lab revolves around the ability to measure cellular events in real time and in living neurons using fluorescence microscopy and electrophysiology. Recent, enormous advances in microscopy have opened amazing new opportunities to measure and to manipulate molecular events as they happen in the cell. To take advantage of this revolution, the lab will continue developing techniques and analysis to understand how actions at the level of molecules and organelles regulate synaptic transmission and other neuronal functions.
Students and postdocs in the lab can expect to utilize state-of-the-art time-lapse microscopy techniques to visualize receptor trafficking and other forms of protein dynamics, along with simultaneous electrophysiological assays of cell surface receptors and channels. Two super-resolution microscopes in the lab are heavily used for single-molecule-based imagine (PALM and STORM). Confocal microscopy is used to perform photobleaching (FRAP), photoactivation, and energy transfer (FRET) analyses. A variety of molecular techniques are used to tag and alter proteins of interest for live-cell imaging, and lab members will be encouraged to develop biochemical assays to test mechanistic hypotheses outside the cell.
Links of Interest
SOM Confocal Microscopy Core Facility
Blanpied Lab web site: https://blanpiedlab.org
